- The central diagonal line through the origin indicates an exact match between the x and y axes as expected. The parallel diagonals off the central line are indicative of repeated sequence elements in different locations of the same protein. Notice the symmetric distribution cross the central diagonal line. For the haptoglobin protein, a repeated sequence is in the regions of 30-90 and 90-150.
- The optimal alignments from two different drosophila olfactory receptors show:
Gap Penalties Identity overlap score -14/-4 20.5% 146aa 100 -5/-1 24.1% 493aa 412 -14/-4 seems more like a local alignment with fewer dispersed gaps and shorter aligned region, whereas -5/-1 seems more like a global alignment with many dispersed gaps and matches. Although -5/-1 looks better on paper, the alignment is full of gaps and not a realistic alignment. Gaps should be placed to allow regions of matching aa's to align.
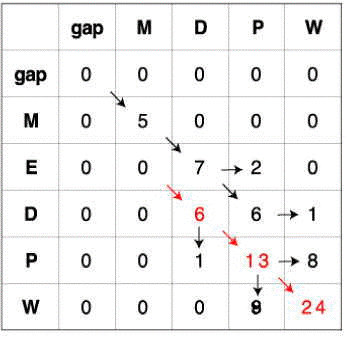
- The steps to do global alignment (with scoring matrix as match +1, mismatch -1, gap -2) are following, reference to the image:
- Insert gaps to the first row and the first column.
- For other cells (each cell is divided into four parts in the image), find the biggest score in each cell among the three (the one with red border):
- insert a gap to the biggest score coming from above (upper right part)
- insert a gap to the biggest score coming from left (bottom left part)
- add the match/mismatch value to the biggest score coming from diagonal (upper left part).
The mathematic calculation for the cell at i-row, j-column: (as shown in parts background color are pink) score = max{SCOREi-1, j + gap, SCOREi,j-1 + gap, SCOREi-1,j-1 + match/mismatch}- remember where the biggest score in each cell coming from.
- Choose the biggest score in the last row and trace back as showing in the following matrix:
The matrix for GAGC and CCG:
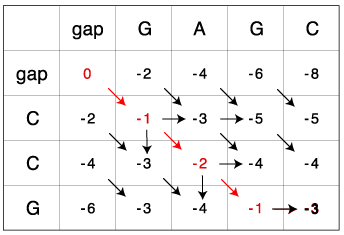
The optimal global alignment from the matrix is:
Adding the individual score based on scoring matrix (match:1, mismatch:-1, gap:-2) according to above alignment:
G A G C C G
(-1) + (-1) + 1 = -1
The above number -1 is identical to the -1 in the bottom row in the matrix. (Look at the image, compare the each part with pink background with its left alignment, this helps you understand why this two number should always be the same)
- The matrix for MDPW and MEDPW is:
The optimal local alignment from the matrix is:
The alignment score is 24.
D P W D P W